Fei-Long Meng’s group and Leng-Siew Yeap’s group shows how DNA flexibility shapes antibody gene hypermutation
Source:Fei-Long Meng
2023-05-30
As the core of the adaptive immune system, diverse antibodies constantly protect the body's health by specifically recognizing and eliminating various pathogens. The process of antibody diversification usually involves two main steps: (1) V(D)J recombination, mediated by the recombinase RAG, and (2) somatic hypermutation (SHM) and class switch recombination (CSR), initiated by the cytidine deaminase AID. SHM introduces high frequency mutations into the antibody variable region. However, the hypermutation is not evenly distributed. Instead, it preferentially accumulates in the complementary determining regions (CDRs) of the variable region. The scientific question of "why mutations have preferences" was first proposed by the famous immunologists David Baltimore and Klaus Rajewsky in 1982. Immunologists have tried to explain this phenomenon from different perspectives for more than 40 years, but have not yet come to a very convincing conclusion. New concepts are therefore urgently needed to solve this classic scientific question of CDR-biased hypermutations.
On April 24, 2023, researchers from the laboratory of Fei-Long Meng at the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences and Leng-Siew Yeap at the Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine have published their research paper entitled "Mesoscale DNA Feature in Antibody-Coding Sequence Facilitates Somatic Hypermutation" in the journal of Cell. This study uncovered the molecular basis of CDR-biased hypermutation and innovatively revealed the important physiological significance of DNA flexibility in the antibody gene somatic hypermutation, resolving a long-standing question that has puzzled antibody researchers for more than 40 years and providing innovative insights into the development of the next-generation antibody humanized animal models.
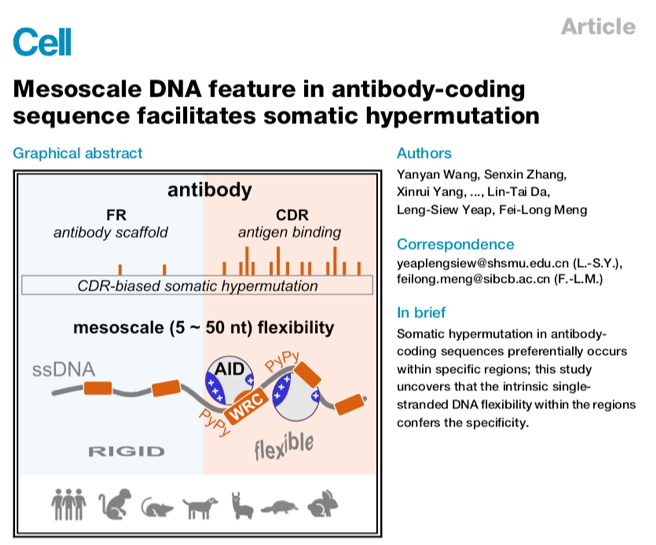
In this study, the researchers established a high-throughput biochemistry assay and found that the simple in vitro enzyme reaction could mimic the CDR-biased hypermutation in vivo, and also found that the CDR-biased hypermutation is evolutionarily conserved in tetrapods that use somatic hypermutation to diversify their antibody repertoire. Taking advantage of the powerful passenger allele mouse model system, which allows the detection of unselected mutational events, and CRISPR/Cas9 system which allows rapid generation of mice with DNA sequence alterations, the researchers found that the mutability of CDR is dependent on the DNA sequence context at the mesoscale level (5-50 bp). Using a combination of molecular dynamics simulations and single-molecule biochemistry, the researchers demonstrated that AID’s targeting preference is directly regulated by the ssDNA flexibility. Analysis of the antibody gene sequence showed that CDR encoding DNA sequences have evolved highly flexible properties. Finally, in the B-cell line and mouse model, inserting flexible DNA into the cold region, FR3, successfully inverted FR into a CDR-like hot region.
This study addresses a classic question that has long plagued the field of immunology. It innovatively points out that there is not only codon-level selection in the evolution of antibody genes, but also a second layer of selection at the non-coding level. The study reveals the crucial regulatory function of DNA flexibility in CDR-biased hypermutation, provides empirical evidence for the regulation of cell life activities by DNA mechanical properties, and suggests that DNA flexibility and other mechanical properties may play important roles in other life activities such as tumor formation and development. This study provides a fundamental theory for the development of next-generation antibody gene humanized animal models.
The study also received a special review on research highlights from the journal Cell Research. The review specifically noted that the study solved an important mystery in the field of somatic hypermutation (SHM) and have broad implications for the evolution of IgV sequences, AID off-target sites, and artificial sequences for the treatment or prevention of viral infections.

Dr. Fei-Long Meng at the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences is the corresponding author of this work. Dr. Leng-Siew Yeap at the Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine is the co-corresponding author. Dr. Yanyan Wang from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences is the first author. This work was supported by National Natural Science Foundation of China, National Key R&D Program of China, and etc.
Links: https://www.cell.com/cell/fulltext/S0092-8674(23)00327-6
On April 24, 2023, researchers from the laboratory of Fei-Long Meng at the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences and Leng-Siew Yeap at the Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine have published their research paper entitled "Mesoscale DNA Feature in Antibody-Coding Sequence Facilitates Somatic Hypermutation" in the journal of Cell. This study uncovered the molecular basis of CDR-biased hypermutation and innovatively revealed the important physiological significance of DNA flexibility in the antibody gene somatic hypermutation, resolving a long-standing question that has puzzled antibody researchers for more than 40 years and providing innovative insights into the development of the next-generation antibody humanized animal models.

In this study, the researchers established a high-throughput biochemistry assay and found that the simple in vitro enzyme reaction could mimic the CDR-biased hypermutation in vivo, and also found that the CDR-biased hypermutation is evolutionarily conserved in tetrapods that use somatic hypermutation to diversify their antibody repertoire. Taking advantage of the powerful passenger allele mouse model system, which allows the detection of unselected mutational events, and CRISPR/Cas9 system which allows rapid generation of mice with DNA sequence alterations, the researchers found that the mutability of CDR is dependent on the DNA sequence context at the mesoscale level (5-50 bp). Using a combination of molecular dynamics simulations and single-molecule biochemistry, the researchers demonstrated that AID’s targeting preference is directly regulated by the ssDNA flexibility. Analysis of the antibody gene sequence showed that CDR encoding DNA sequences have evolved highly flexible properties. Finally, in the B-cell line and mouse model, inserting flexible DNA into the cold region, FR3, successfully inverted FR into a CDR-like hot region.
This study addresses a classic question that has long plagued the field of immunology. It innovatively points out that there is not only codon-level selection in the evolution of antibody genes, but also a second layer of selection at the non-coding level. The study reveals the crucial regulatory function of DNA flexibility in CDR-biased hypermutation, provides empirical evidence for the regulation of cell life activities by DNA mechanical properties, and suggests that DNA flexibility and other mechanical properties may play important roles in other life activities such as tumor formation and development. This study provides a fundamental theory for the development of next-generation antibody gene humanized animal models.
The study also received a special review on research highlights from the journal Cell Research. The review specifically noted that the study solved an important mystery in the field of somatic hypermutation (SHM) and have broad implications for the evolution of IgV sequences, AID off-target sites, and artificial sequences for the treatment or prevention of viral infections.

Dr. Fei-Long Meng at the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences is the corresponding author of this work. Dr. Leng-Siew Yeap at the Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine is the co-corresponding author. Dr. Yanyan Wang from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences is the first author. This work was supported by National Natural Science Foundation of China, National Key R&D Program of China, and etc.
Links: https://www.cell.com/cell/fulltext/S0092-8674(23)00327-6


