Shuo Wang' s Team Reveals Epigenetic Imprints Driving Lineage Segregation of Mucosal Innate Lymphoid Cells
Source:Shuo Wang
2025-10-16
The immune system is established through the hierarchical development and differentiation of immune cells—a process meticulously regulated by a multitude of internal and external factors. Recent research has uncovered heterogeneity among immune progenitors, such as hematopoietic stem cells. These progenitors exhibit a predisposition to differentiate into particular types of immune effector cells. However, the factors governing this lineage bias remain poorly defined, and the regulatory mechanisms that steer immune precursors toward distinct effector fates warrant further investigation.
Innate lymphoid cells (ILCs) are a subset of innate lymphocytes enriched in mucosal tissues and functionally analogous to T helper (Th) cells in many aspects. Despite these functional parallels, their developmental pathways diverge considerably. ILC precursors (ILCPs) develop in the bone marrow, in contrast to T cell precursors, which mature in the thymus. In peripheral tissues, dendritic cells (DCs) present antigens and secrete cytokines to drive the differentiation of specialized Th cell subsets. Conversely, ILC maturation and differentiation are antigen-independent, yet diverse ILC subsets still emerge within similar mucosal microenvironments. This fundamental difference suggests that the differentiation and fate commitment of distinct ILC subsets are likely governed by unique regulatory programs, a central and unresolved question in the field.
The research group led by Professor Shuo Wang at the Institute of Microbiology, Chinese Academy of Sciences (CAS), focuses on the establishment and homeostatic regulation of the mucosal immune system. Their work has elucidated the functional mechanisms by which the gut microbiota regulates mucosal immune homeostasis through epigenetic modifications. Building on this foundation, the team further investigated the epigenetic imprinting during the development of mucosal innate lymphoid cells (ILCs). On Sep 18, 2025, they published a research paper in Nature Immunology titled “Epigenetic imprinting in innate lymphoid cell precursors directs the lineage segregation of innate lymphoid cells”. This study systematically reveals the critical role of epigenetic imprints in innate lymphoid cell precursors (ILCPs) in determining lineage differentiation, establishes a DNA methylation-based tracing model, and provides new insights into the epigenetic regulation of mucosal immune system development and immune cell differentiation.
To investigate the heterogeneity of ILCPs and the changes in transcriptomic and epigenetic modifications during their differentiation into various ILC subsets, the researchers generated single-cell profiles of DNA methylation, chromatin accessibility, and transcriptomes for ILCPs and ILC subsets. They identified two distinct ILCP subpopulations: ILCP1 and ILCP2. ILCP2s exhibited lower overall DNA methylation levels compared to ILCP1s and displayed highly heterogeneous methylation patterns. Further dimensionality reduction and clustering analysis revealed that ILCP2s could be categorized into three subtypes—ILCP2-ILC1, ILCP2-ILC2, and ILCP2-ILC3—based on their DNA methylation characteristics. These ILCP2 subtypes showed methylation signatures corresponding to their respective effector ILC lineages.
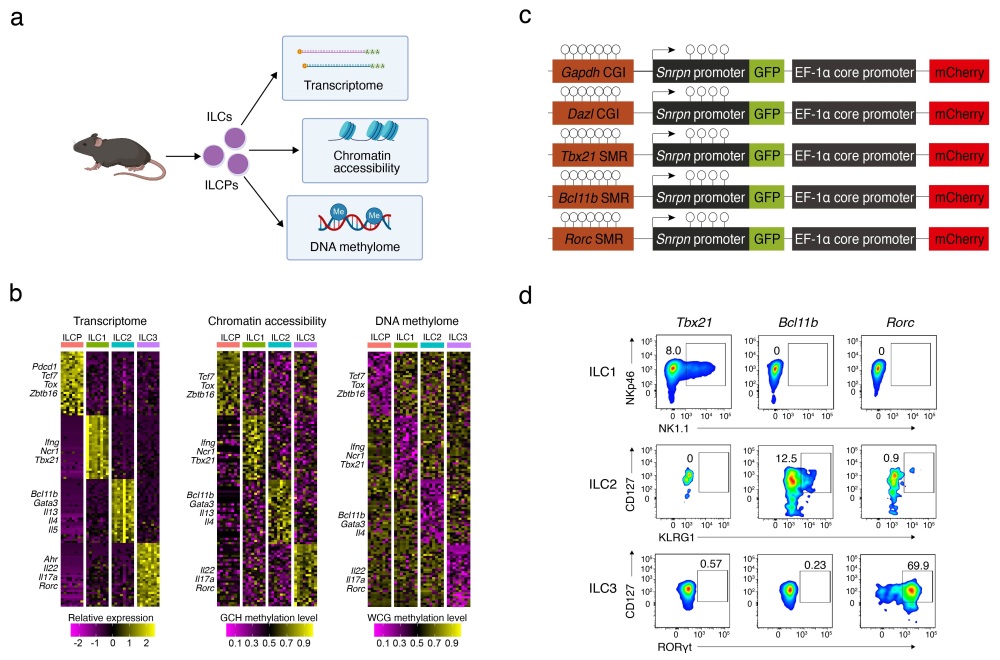
The researchers further defined and characterized signature methylation regions (SMRs) for each ILCP2 subtype and developed an SMR-based lineage tracing technique to track DNA methylation imprints during ILCP differentiation. They found that ILCP2s carrying hypomethylated SMRs specific to a particular ILC lineage preferentially differentiated into that corresponding ILC subset. DNA methylation editing of SMRs suppressed ILC lineage differentiation, and knockout of the DNA methyltransferase Dnmt1 gene in ILCPs disrupted the heterogeneous distribution of SMRs and led to defective ILC differentiation. These results demonstrate that ILCPs exhibit heterogeneous DNA methylation patterns with lineage-specific features, and that epigenetic imprints in the form of DNA methylation drive the directed lineage differentiation of ILCs during development. This study highlights the decisive role of epigenetic imprinting in the functional establishment of mucosal ILCs and provides a new theoretical framework for understanding the regulation of immune cell fate determination.
The co-first authors of the study are Drs. Zhen Liu, Fei Shao from the Institute of Microbiology, CAS, and Dr. Qiang Zhang from Xiamen University. Corresponding author is Prof. Shuo Wang from the Institute of Microbiology, CAS. The research received support and contributions from Prof. Shuai Gao (China Agricultural University), Prof. Pengyan Xia (Peking University), and Prof. Zhuqiang Zhang (Institute of Biophysics, CAS). This work was supported by the National Key R&D Program of China, the Beijing Natural Science Foundation, and the Key Research Program of Frontier Sciences of CAS.
Article Link: https://www.nature.com/articles/s41590-025-02261-0
Innate lymphoid cells (ILCs) are a subset of innate lymphocytes enriched in mucosal tissues and functionally analogous to T helper (Th) cells in many aspects. Despite these functional parallels, their developmental pathways diverge considerably. ILC precursors (ILCPs) develop in the bone marrow, in contrast to T cell precursors, which mature in the thymus. In peripheral tissues, dendritic cells (DCs) present antigens and secrete cytokines to drive the differentiation of specialized Th cell subsets. Conversely, ILC maturation and differentiation are antigen-independent, yet diverse ILC subsets still emerge within similar mucosal microenvironments. This fundamental difference suggests that the differentiation and fate commitment of distinct ILC subsets are likely governed by unique regulatory programs, a central and unresolved question in the field.
The research group led by Professor Shuo Wang at the Institute of Microbiology, Chinese Academy of Sciences (CAS), focuses on the establishment and homeostatic regulation of the mucosal immune system. Their work has elucidated the functional mechanisms by which the gut microbiota regulates mucosal immune homeostasis through epigenetic modifications. Building on this foundation, the team further investigated the epigenetic imprinting during the development of mucosal innate lymphoid cells (ILCs). On Sep 18, 2025, they published a research paper in Nature Immunology titled “Epigenetic imprinting in innate lymphoid cell precursors directs the lineage segregation of innate lymphoid cells”. This study systematically reveals the critical role of epigenetic imprints in innate lymphoid cell precursors (ILCPs) in determining lineage differentiation, establishes a DNA methylation-based tracing model, and provides new insights into the epigenetic regulation of mucosal immune system development and immune cell differentiation.
To investigate the heterogeneity of ILCPs and the changes in transcriptomic and epigenetic modifications during their differentiation into various ILC subsets, the researchers generated single-cell profiles of DNA methylation, chromatin accessibility, and transcriptomes for ILCPs and ILC subsets. They identified two distinct ILCP subpopulations: ILCP1 and ILCP2. ILCP2s exhibited lower overall DNA methylation levels compared to ILCP1s and displayed highly heterogeneous methylation patterns. Further dimensionality reduction and clustering analysis revealed that ILCP2s could be categorized into three subtypes—ILCP2-ILC1, ILCP2-ILC2, and ILCP2-ILC3—based on their DNA methylation characteristics. These ILCP2 subtypes showed methylation signatures corresponding to their respective effector ILC lineages.

The researchers further defined and characterized signature methylation regions (SMRs) for each ILCP2 subtype and developed an SMR-based lineage tracing technique to track DNA methylation imprints during ILCP differentiation. They found that ILCP2s carrying hypomethylated SMRs specific to a particular ILC lineage preferentially differentiated into that corresponding ILC subset. DNA methylation editing of SMRs suppressed ILC lineage differentiation, and knockout of the DNA methyltransferase Dnmt1 gene in ILCPs disrupted the heterogeneous distribution of SMRs and led to defective ILC differentiation. These results demonstrate that ILCPs exhibit heterogeneous DNA methylation patterns with lineage-specific features, and that epigenetic imprints in the form of DNA methylation drive the directed lineage differentiation of ILCs during development. This study highlights the decisive role of epigenetic imprinting in the functional establishment of mucosal ILCs and provides a new theoretical framework for understanding the regulation of immune cell fate determination.
The co-first authors of the study are Drs. Zhen Liu, Fei Shao from the Institute of Microbiology, CAS, and Dr. Qiang Zhang from Xiamen University. Corresponding author is Prof. Shuo Wang from the Institute of Microbiology, CAS. The research received support and contributions from Prof. Shuai Gao (China Agricultural University), Prof. Pengyan Xia (Peking University), and Prof. Zhuqiang Zhang (Institute of Biophysics, CAS). This work was supported by the National Key R&D Program of China, the Beijing Natural Science Foundation, and the Key Research Program of Frontier Sciences of CAS.
Article Link: https://www.nature.com/articles/s41590-025-02261-0


