Mapping 10 Million Immune Cells to Decode Disease Mechanisms: New Atlas and AI Model Pave Way for Precision Immunotherapy
Source:Tingting Wen
2026-01-13
On January 8, 2026, a multi-institutional research team led by the State Key Laborato-ry of Genome and Multi-omics Technologies (initiated by BGI-Research), working with clinical and academic partners including Ruijin Hospital affiliated with Shanghai Jiao Tong University School of Medicine, Shanxi Medical University, and other collaborators, reports in Science the Chinese Immune Multi-Omics Atlas (CIMA).
By integrating single-cell transcriptomics, single-cell chromatin accessibility, whole-genome sequencing, and plasma metabolomic, lipidomic, and clinical biochemistry profiles, the study offers a high-resolution reference for how age, sex, and genetic var-iation shape circulating immune cells in adults. The resource addresses a long-standing bottleneck in large-scale single-cell epigenomic data and expands population representation in mechanistic immune studies.
At the heart of CIMA is a cohort of 428 adults aged 20 to 77 years who self-reported no active disease at the time of sampling. The team profiled more than 10 million pe-ripheral blood immune cells, retaining 6.5 million high-quality cells from single-cell RNA sequencing and 3.8 million from single-cell ATAC sequencing after quality control. This scale enabled the identification of 73 immune cell types, including rare populations be-low 0.1% frequency, and revealed immune cell subsets and molecular features that vary with age and sex.
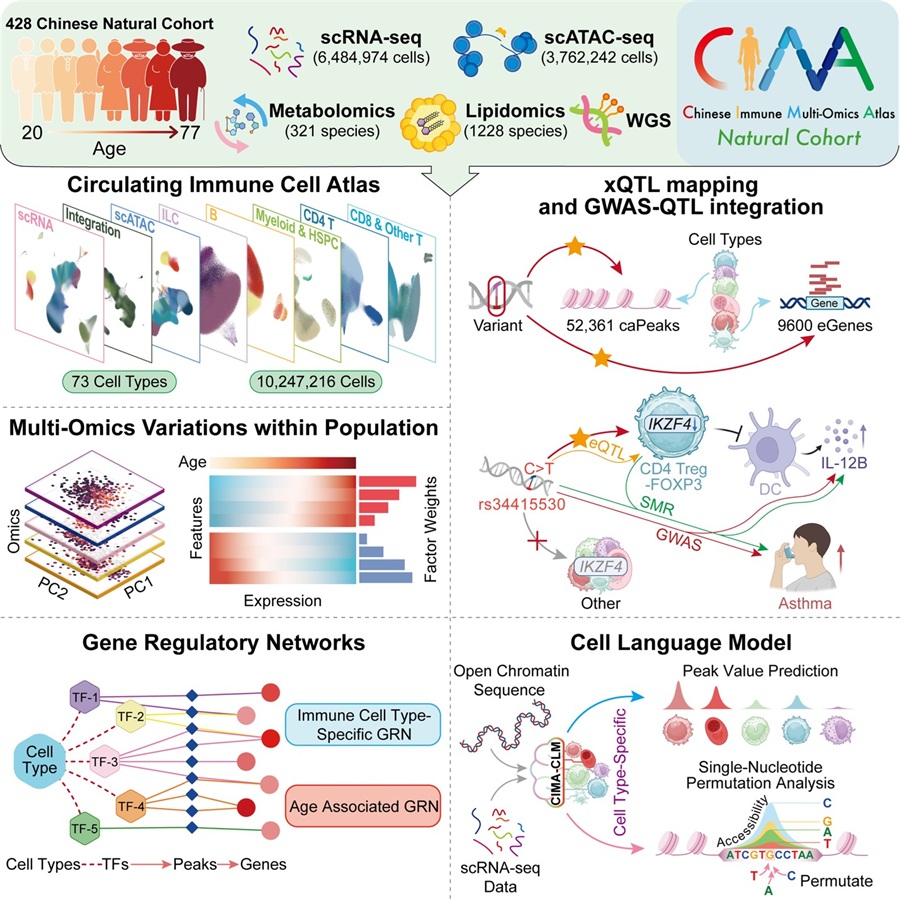
Using paired gene expression and chromatin accessibility data, the researchers con-structed enhancer-driven gene regulatory networks that capture how transcription fac-tors coordinate immune cell identity. They identified 404 enhancer-linked regulatory units, comprising 84,625 regulatory regions and 13,645 target genes, and systemati-cally mapped the regulatory relationships among key transcription factors, cis-regulatory elements, and target genes across 61 immune cell subtypes.
To connect regulatory variation to inherited genetics, the team performed whole-genome sequencing and mapped cell type-resolved expression quantitative trait loci (eQTLs) and chromatin accessibility QTLs (caQTLs). The analysis identified 9,600 eGenes and 52,361 caPeaks, with substantial cell type specificity—nearly 30% of eGenes and 55% of caPeaks were specific to a single cell type. This reinforces that many disease-relevant genetic effects are not "blood-wide" but concentrated within particular immune subtypes.
A key demonstration of the atlas's mechanistic value comes from integrative analyses linking variants to molecular traits and disease risk. Using summary-data-based Men-delian randomization across 154 traits, including plasma lipids, metabolites, inflamma-tory proteins, and immune-related diseases, the researchers identified 1,196 signifi-cant pleiotropic associations across 68 immune cell types. One illustrative example in-volves rs34415530, which is associated with cell-specific regulation of IKZF4 in CD4 Treg-FOXP3 cells and with circulating IL-12B protein levels and asthma susceptibility. These findings provide biologically grounded hypotheses for where and how noncod-ing risk variants may shape immune-mediated disease processes.
Beyond the atlas itself, the study introduces CIMA-CLM, a cell language model that integrates chromatin sequence features and single-cell gene expression to predict chromatin accessibility and assess the functional impact of noncoding variants. Across 32 immune cell types, CIMA-CLM achieved high concordance with experimental pro-files, with an overall mean Pearson correlation of 0.8951 and a mean AUROC of 0.9560. By enabling in silico mutational analyses, the model offers a new computa-tional route to explore the regulatory consequences of disease-associated variants.
This research framework also demonstrates the potential to link and integrate cell atlas analysis with more general genome-based foundational models (such as Genos). This fusion aims to build a multi-level, interpretable intelligent prediction framework from DNA sequences to cellular functions, paving the way for understanding life regulatory mechanisms and accelerating biomedical discoveries.
This study is a successful demonstration of BGI Group's "133111i" Multi-Omics Preci-sion Health Management System, a new paradigm for precision health research. By integrating multi-dimensional information such as single-cell transcriptomics, epige-nomics, plasma metabolomics, lipidomic, and physiological characteristics, CIMA es-tablished a multi-omics baseline for immune health. It enables the digital analysis and precise insights into complex life systems, empowering disease prevention and health management.
This systematic research framework and the high-quality data it generated have al-ready driven innovation incubation. Based on this, a BGI AI evaluation model for per-sonal immune capability was developed, enabling high-precision and comprehensive assessments of individual immune states. This achievement marks the effective trans-lation of cutting-edge scientific research into accessible healthcare service.
CIMA also serves as a reference for the international immunology and genomics communities. By anchoring genetic effects in specific cell types and regulatory ele-ments, and by providing open data and interactive tools through the CIMA Portal, the atlas helps researchers refine the biological interpretation of GWAS signals, compare immune regulatory architecture across ancestries, and explore how age and sex inter-act with genetic regulation in blood.
As noted by Tao Cheng, Academician of the Chinese Academy of Engineering and Director of the Institute of Hematology at the Chinese Academy of Medical Sciences, and Guang Ning, Academician of the Chinese Academy of Engineering at Ruijin Hos-pital affiliated with Shanghai Jiao Tong University School of Medicine, CIMA's cell type-specific regulatory networks provide crucial insights for immune-metabolic interactions in diseases such as atherosclerosis and type 2 diabetes, and integration with other Chinese population cohorts will advance precision medicine for complex diseases.
Building on this foundation, the larger-scale Phase II CIMA initiative has officially launched. Its research scope will expand from healthy populations to major disease cohorts, including autoimmune diseases, cardiovascular diseases, and infectious dis-eases. By leveraging advanced technologies such as Stereo-cell and protein multiplex detection, the initiative aims to systematically unravel the immunological mechanisms underlying disease onset and progression, identify new diagnostic and therapeutic tar-gets, and provide high-quality data resources for constructing more precise "virtual cell" models, enabling digital prediction of disease simulation and intervention strate-gies.
All participants provided written informed consent, and candidates with active disease and pregnant women were excluded. Ethics approval for this study is obtained.
Article link: https://www.science.org/doi/10.1126/science.adt3130
By integrating single-cell transcriptomics, single-cell chromatin accessibility, whole-genome sequencing, and plasma metabolomic, lipidomic, and clinical biochemistry profiles, the study offers a high-resolution reference for how age, sex, and genetic var-iation shape circulating immune cells in adults. The resource addresses a long-standing bottleneck in large-scale single-cell epigenomic data and expands population representation in mechanistic immune studies.
At the heart of CIMA is a cohort of 428 adults aged 20 to 77 years who self-reported no active disease at the time of sampling. The team profiled more than 10 million pe-ripheral blood immune cells, retaining 6.5 million high-quality cells from single-cell RNA sequencing and 3.8 million from single-cell ATAC sequencing after quality control. This scale enabled the identification of 73 immune cell types, including rare populations be-low 0.1% frequency, and revealed immune cell subsets and molecular features that vary with age and sex.

Mapping the Chinese Immune Landscape. By integrating multi-omics data from 428 individuals to catego-rize over 10 million immune cells, this study establishes a high-definition reference atlas for East Asian immune diversity, providing a critical baseline for precision medicine.
Using paired gene expression and chromatin accessibility data, the researchers con-structed enhancer-driven gene regulatory networks that capture how transcription fac-tors coordinate immune cell identity. They identified 404 enhancer-linked regulatory units, comprising 84,625 regulatory regions and 13,645 target genes, and systemati-cally mapped the regulatory relationships among key transcription factors, cis-regulatory elements, and target genes across 61 immune cell subtypes.
To connect regulatory variation to inherited genetics, the team performed whole-genome sequencing and mapped cell type-resolved expression quantitative trait loci (eQTLs) and chromatin accessibility QTLs (caQTLs). The analysis identified 9,600 eGenes and 52,361 caPeaks, with substantial cell type specificity—nearly 30% of eGenes and 55% of caPeaks were specific to a single cell type. This reinforces that many disease-relevant genetic effects are not "blood-wide" but concentrated within particular immune subtypes.
A key demonstration of the atlas's mechanistic value comes from integrative analyses linking variants to molecular traits and disease risk. Using summary-data-based Men-delian randomization across 154 traits, including plasma lipids, metabolites, inflamma-tory proteins, and immune-related diseases, the researchers identified 1,196 signifi-cant pleiotropic associations across 68 immune cell types. One illustrative example in-volves rs34415530, which is associated with cell-specific regulation of IKZF4 in CD4 Treg-FOXP3 cells and with circulating IL-12B protein levels and asthma susceptibility. These findings provide biologically grounded hypotheses for where and how noncod-ing risk variants may shape immune-mediated disease processes.
Beyond the atlas itself, the study introduces CIMA-CLM, a cell language model that integrates chromatin sequence features and single-cell gene expression to predict chromatin accessibility and assess the functional impact of noncoding variants. Across 32 immune cell types, CIMA-CLM achieved high concordance with experimental pro-files, with an overall mean Pearson correlation of 0.8951 and a mean AUROC of 0.9560. By enabling in silico mutational analyses, the model offers a new computa-tional route to explore the regulatory consequences of disease-associated variants.
This research framework also demonstrates the potential to link and integrate cell atlas analysis with more general genome-based foundational models (such as Genos). This fusion aims to build a multi-level, interpretable intelligent prediction framework from DNA sequences to cellular functions, paving the way for understanding life regulatory mechanisms and accelerating biomedical discoveries.
This study is a successful demonstration of BGI Group's "133111i" Multi-Omics Preci-sion Health Management System, a new paradigm for precision health research. By integrating multi-dimensional information such as single-cell transcriptomics, epige-nomics, plasma metabolomics, lipidomic, and physiological characteristics, CIMA es-tablished a multi-omics baseline for immune health. It enables the digital analysis and precise insights into complex life systems, empowering disease prevention and health management.
This systematic research framework and the high-quality data it generated have al-ready driven innovation incubation. Based on this, a BGI AI evaluation model for per-sonal immune capability was developed, enabling high-precision and comprehensive assessments of individual immune states. This achievement marks the effective trans-lation of cutting-edge scientific research into accessible healthcare service.
CIMA also serves as a reference for the international immunology and genomics communities. By anchoring genetic effects in specific cell types and regulatory ele-ments, and by providing open data and interactive tools through the CIMA Portal, the atlas helps researchers refine the biological interpretation of GWAS signals, compare immune regulatory architecture across ancestries, and explore how age and sex inter-act with genetic regulation in blood.
As noted by Tao Cheng, Academician of the Chinese Academy of Engineering and Director of the Institute of Hematology at the Chinese Academy of Medical Sciences, and Guang Ning, Academician of the Chinese Academy of Engineering at Ruijin Hos-pital affiliated with Shanghai Jiao Tong University School of Medicine, CIMA's cell type-specific regulatory networks provide crucial insights for immune-metabolic interactions in diseases such as atherosclerosis and type 2 diabetes, and integration with other Chinese population cohorts will advance precision medicine for complex diseases.
Building on this foundation, the larger-scale Phase II CIMA initiative has officially launched. Its research scope will expand from healthy populations to major disease cohorts, including autoimmune diseases, cardiovascular diseases, and infectious dis-eases. By leveraging advanced technologies such as Stereo-cell and protein multiplex detection, the initiative aims to systematically unravel the immunological mechanisms underlying disease onset and progression, identify new diagnostic and therapeutic tar-gets, and provide high-quality data resources for constructing more precise "virtual cell" models, enabling digital prediction of disease simulation and intervention strate-gies.
All participants provided written informed consent, and candidates with active disease and pregnant women were excluded. Ethics approval for this study is obtained.
Article link: https://www.science.org/doi/10.1126/science.adt3130


